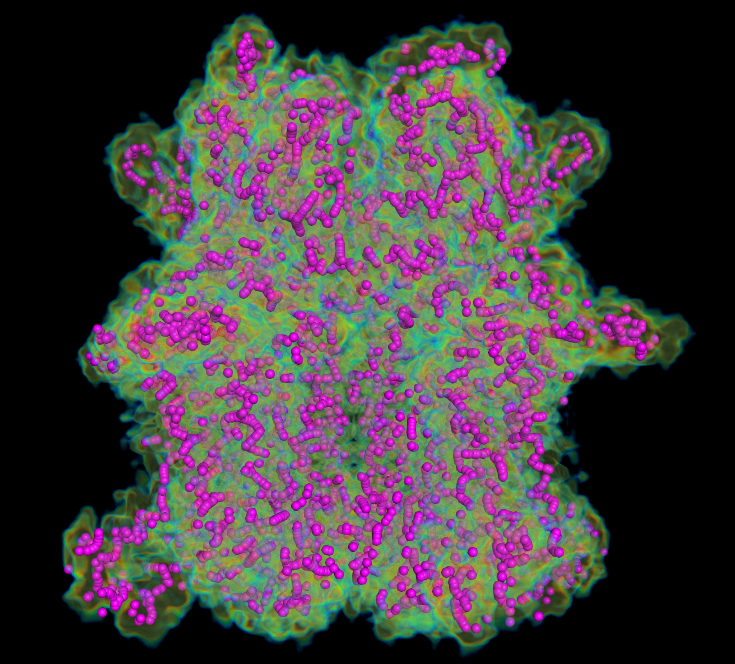
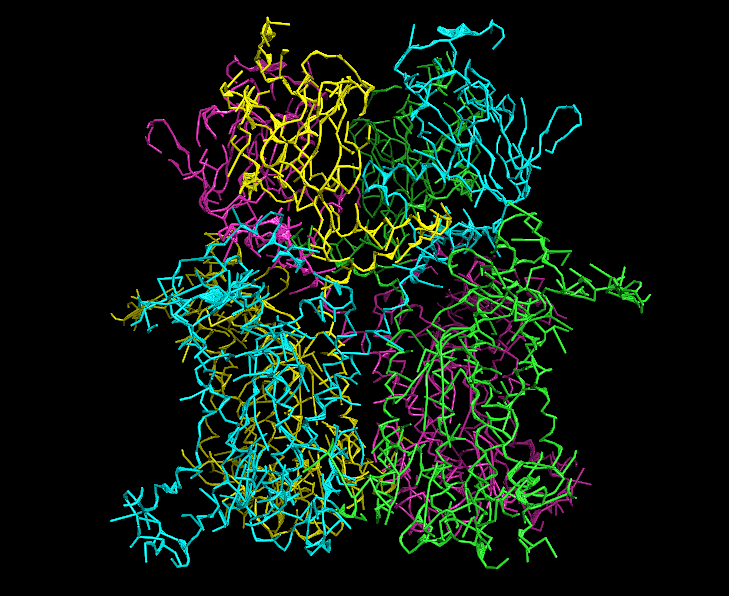
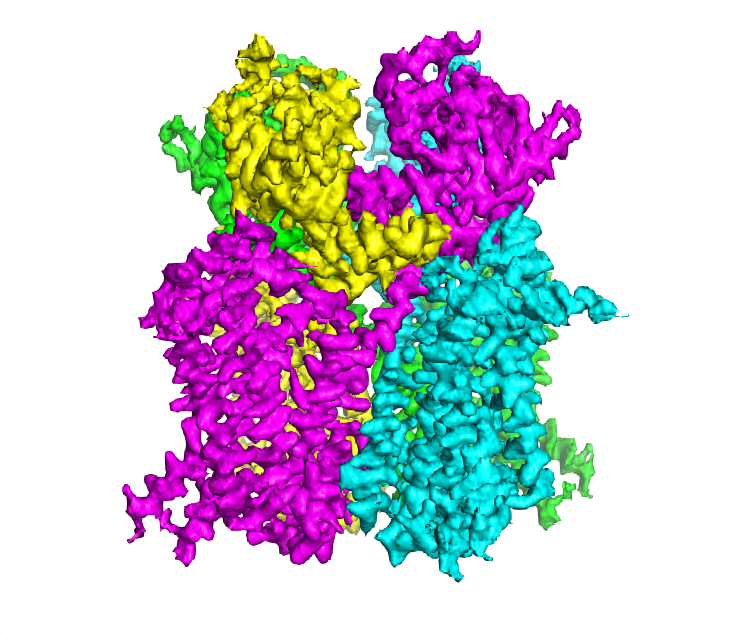
MAINMASTseg
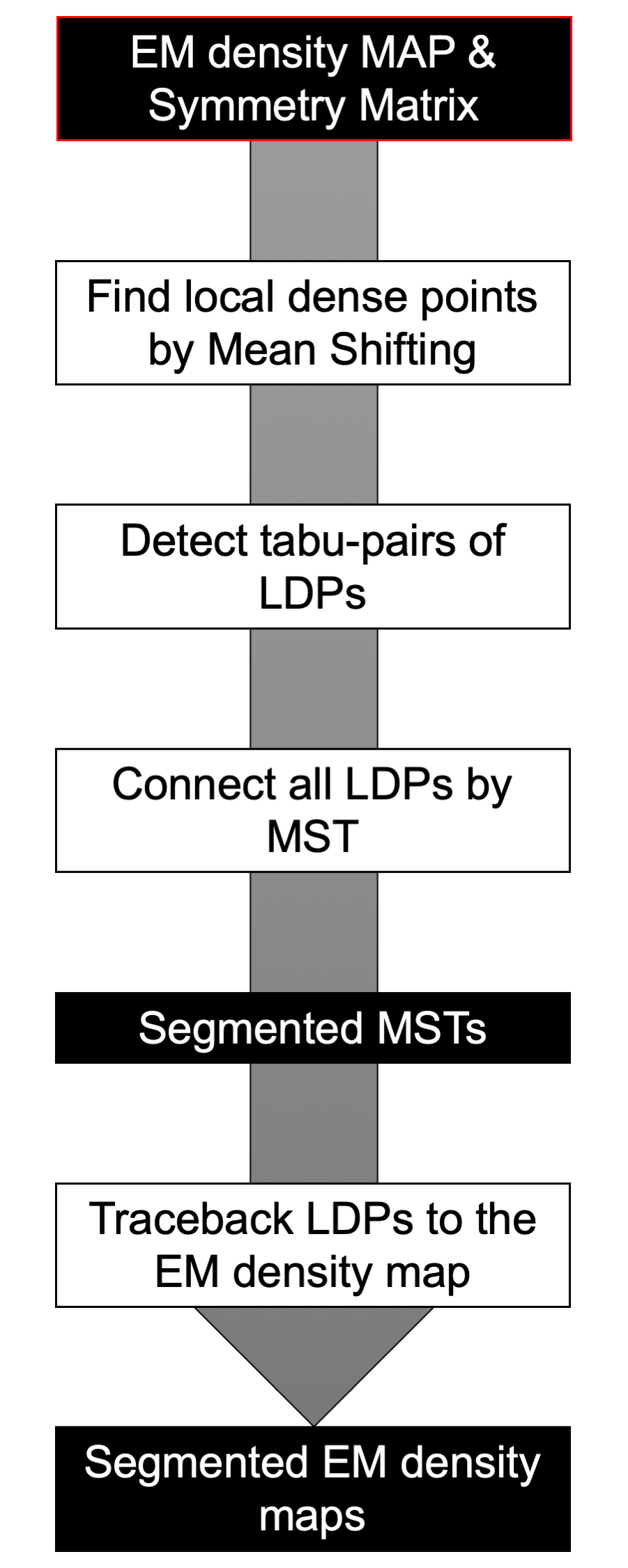
Mainm program. MAINMASTseg computes segmented density maps (mrc format) and builds MST model in CIF format.
Usage: MainmastSeg -i [MAP.mrc] -Y [Rotation Matrix] [(option)] Fast MainmastSeg C-lang & multi thread version ---Mode--- -L : Fast LDP search mode -M : Minimum Spanning Tree Mode (default) -G : Graph Mode -W : Generating MRC file Mode. File name segment%d.mrc -V : Movie Mode ---Options--- -c [int ] :Number of cores for threads def=2 -t [float] :Threshold of density map def=0.000 -g [float] : bandwidth of the gaussian filter def=2.0, sigma = 0.5*[float] -f [float] :Filtering for representative points def=0.100 -m [float] :After MeanShifting merge LDPs where d<[float] def=0.500 -R [float] :Radius of Local MST def=10.000 -k [float] :keep edges where d < [float] def=0.500 Thi is Ver 1.041

 MAINMASTseg protocol
MAINMASTseg protocol