EM-GAN
EM-GAN is a computational tool, which enables capturing protein structure information from cryo-EM maps more effectively than raw maps. It is based on 3D deep learning. It is aimed to help protein structure modeling from cryo-EM maps.
Introduction
EM-GAN uses a 3D deep neural network architecture, Generative Adversarial Networks (GAN), to modify an input cryo-EM map of 3 Å to 6 Å to enable more efficient extraction of protein structure information from the input map. In our benchmark, protein structure modeling tools were able to build more accurate protein structure models by using output from EM-GAN. EM-GAN takes local regions (patches) of experimental EM map and processed patches are then combined together to output a whole EM map.
GAN architecture of EM-GAN
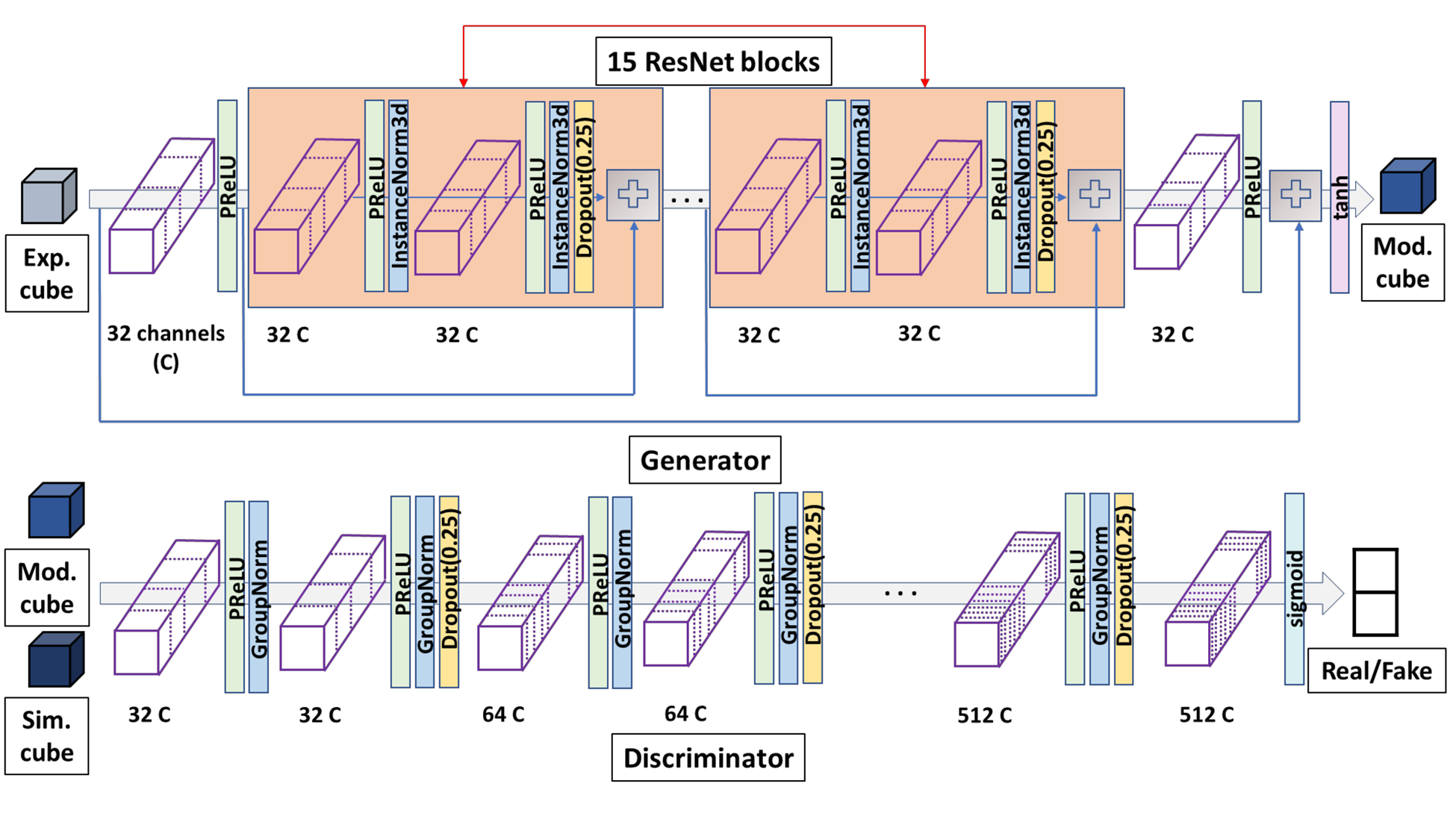
Figure 1. The GAN architecture of EM-GAN. The detailed architecture of the generator and discriminator networks are shown. The blue arrows that connect the input of a ResNet block and a plus sign, which is the operator that simply add two matrices, is a skip connection.
Tutorial
Architecture
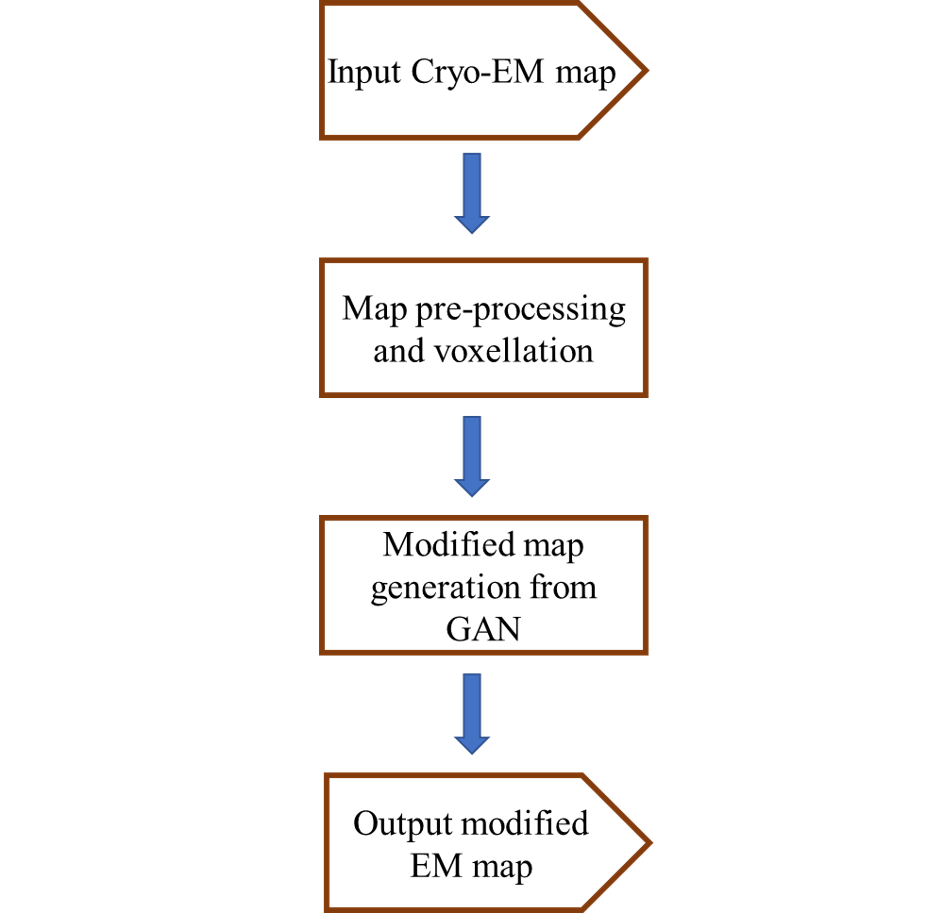
The architecture of EM-GAN is summarized in the flowchart on the right.
This document will provide a detailed explanation of each step of the EM-GAN architecture and programs needed to run those steps. It concludes by giving a step-by-step application walk-through on an example EM map.
Input and pre-processing of your EM map
The input file for EM-GAN is generated in two-steps. In the first step, a program named HLmapData_new takes an EM map and relevant details such as the map's author recommended contour level and pixels/Angstrom values as input and generates an intermediate readable text file called [your_map_id]_trimmap. A trimmap contains normalized electron density values of voxels. In the second step, a program named dataset uses this trimmap to generate an input file called [your_map_id]_dataset, which contains rows of density values that we get by scanning the input map in all the 3 directions using a 25*25*25 cube.
EM-GAN modified map generation
The test dataset generated in the previous step is fed to the EM-GAN architecture that contains a Generative Adversarial Network (GAN). This step outputs 25*25*25 Å^3 modified cube for every corresponding input cube of similar dimensions.
Usage guide
For detailed step by step usage guide, please visit here.
Examples
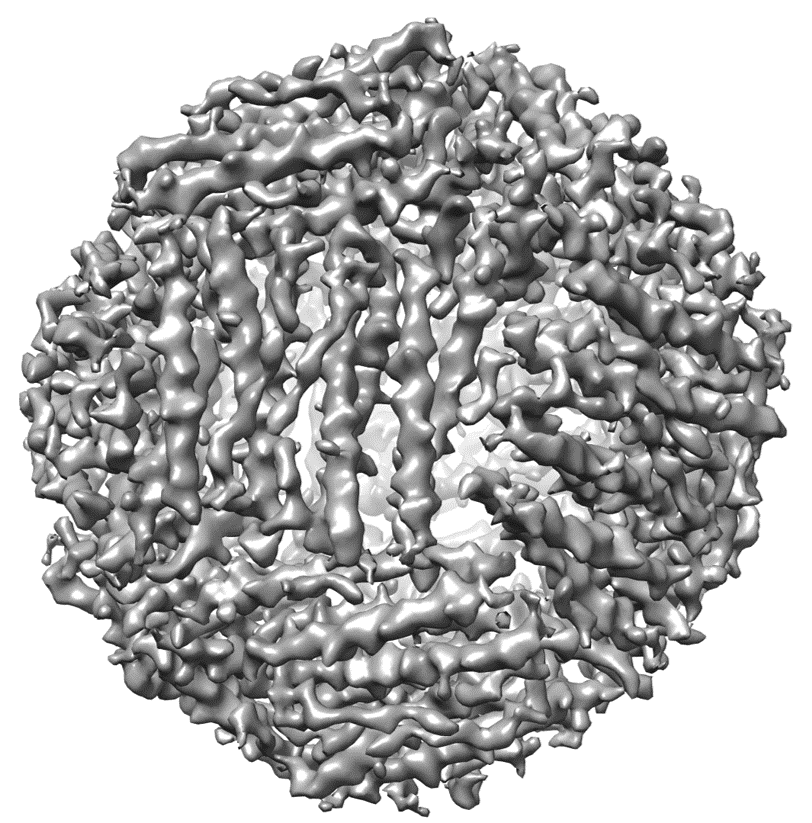
Experimental map example (EMD-2788)
You can download the EM map for protein structure with EMID 2788 here. Use this map file and follow the instructions in step 1 of usage guide to generate input dataset file.The trimmap file is generated as
data_prep/HLmapData_new 2788.mrc -c 0.16 > 2788_trimmap
The author recommended contour level for the map EMD-2788 is 0.16 which has been provided as one of the options above.
You can generate the input dataset file as follows,
python data_prep/dataset_final.py 2788_trimmap 2788_dataset 2788
If the generated input file is 2788_dataset, write the file location to a dataset location file as follows
echo ./2788_dataset > test_dataset_location
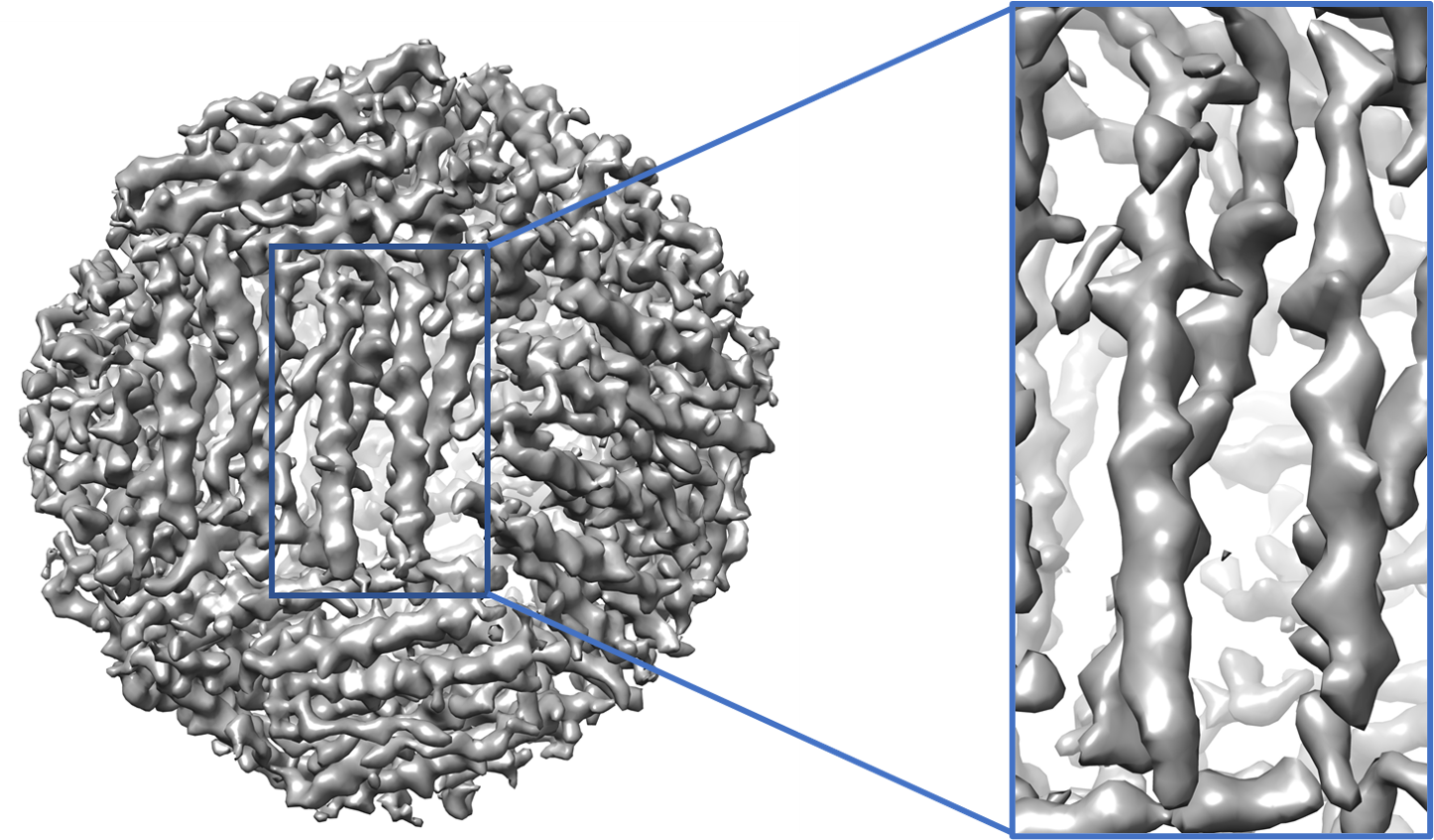
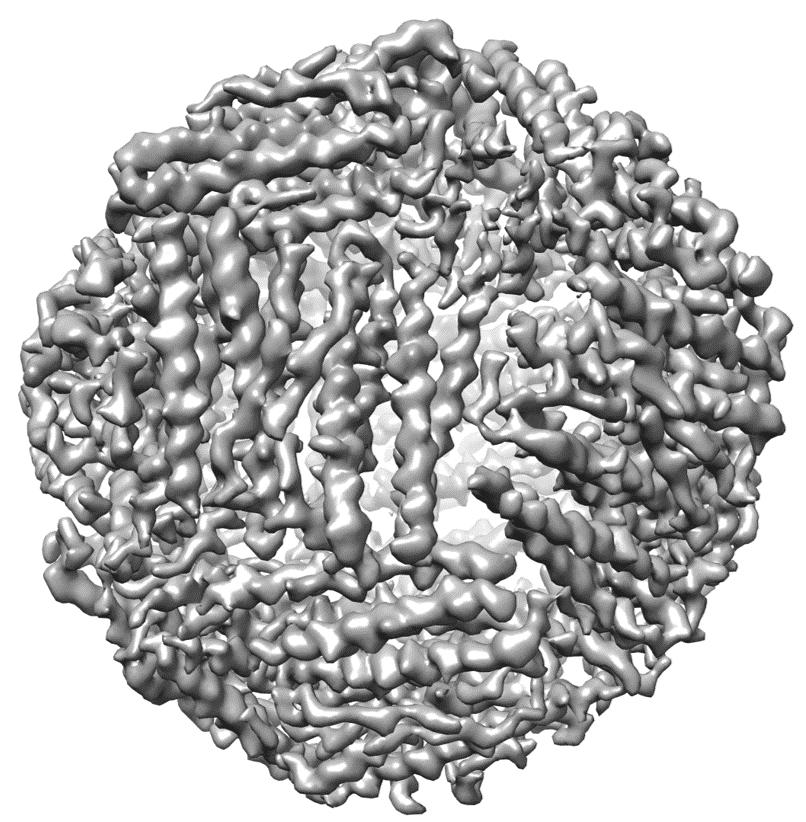
EM-GAN modified map generation
You can then run the EM-GAN program to generate modified EM map as follows
python test.py --input=test_dataset_location --G_res_blocks=15 --D_res_blocks=3 --G_path=model/Generator --D_path=model/Generator
An example of generated map of EMD-2788 is shown on the right.
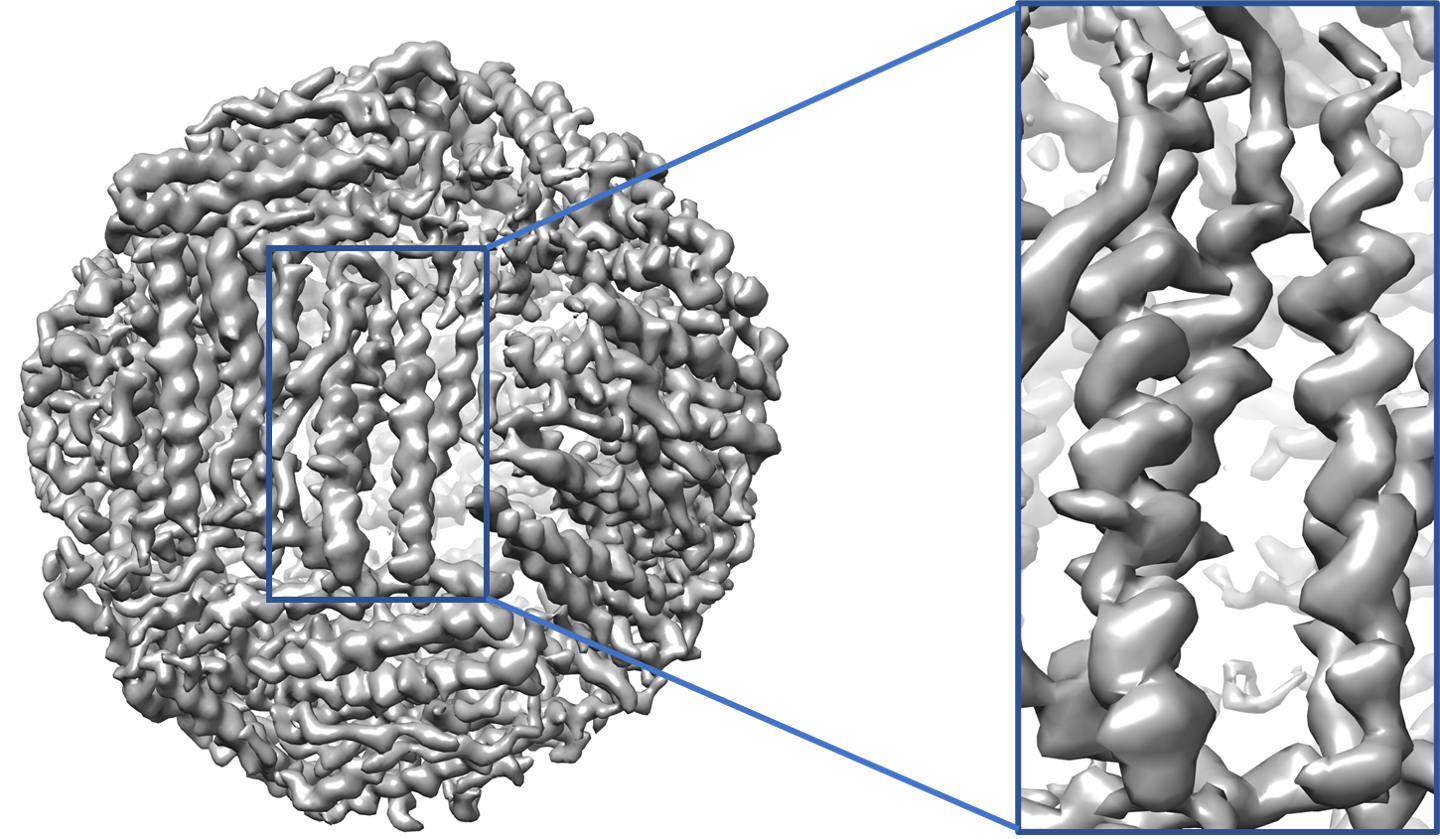
Availability
EM-GAN on GoogleColab is available at EM-GAN Google Colab version
Tech Specs
CPU: >=4 cores
Memory: >=10Gb
GPU: any GPU supports CUDA with more than 12GB memory.
License
Copyright © 2020 Sai Raghavendra Maddhuri Venkata Subramaniya, Genki Terashi, Daisuke Kihara, and Purdue University.
EM-GAN is a free software for academic and non-commercial users.
It is released under the terms of the GNU General Public License Ver.3 (https://www.gnu.org/licenses/gpl-3.0.en.html).
Commercial users please contact dkihara@purdue.edu for alternate licensing.
Reference
Citation of the following reference should be included in any publication that uses data or results generated by EM-GAN program.
