MainMast FAQ
Installation
Q. I want to use MAINMAST program on MacOSX. But I can not use gfortran.
A. To use gfortran, you have to install Xcode, Homebrew on your Mac.
Open App Store. See Xcode
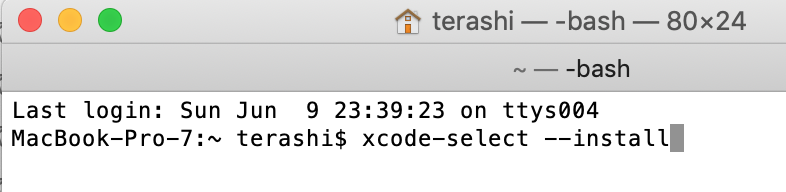
Open "terminal" from Applications, then
Type command
xcode-select --install
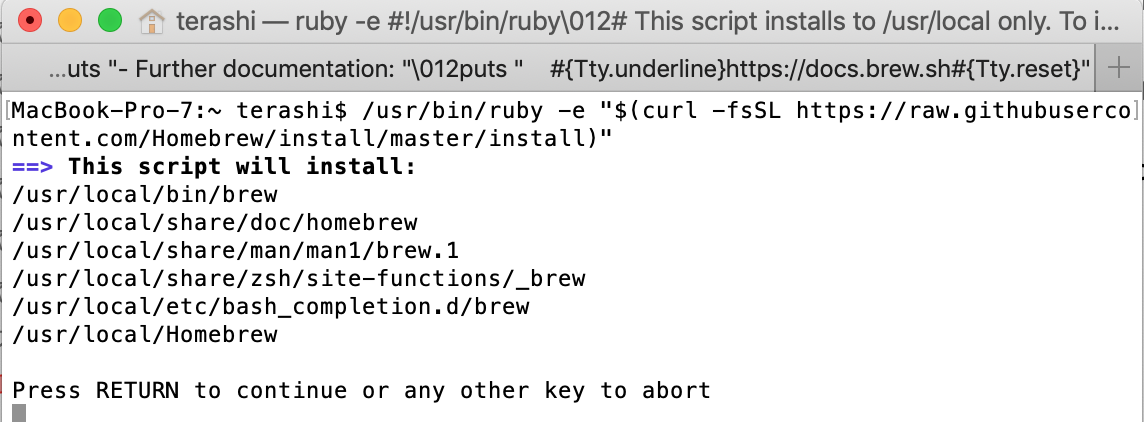
Type command in terminal window
/usr/bin/ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)"
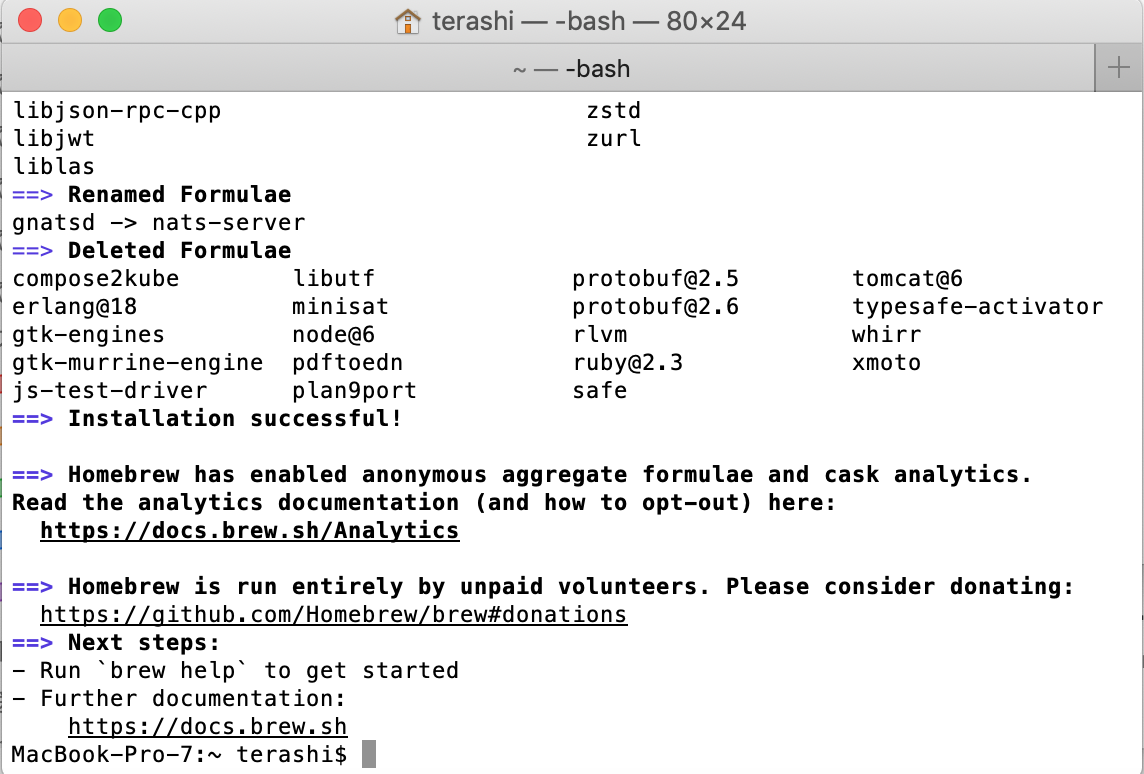
Confirm the homebrew command
brew doctor
Type command in terminal window
brew install gcc
Confirm the gfortran command.
gfortran -v
Input Data issues
Q. MAINMAST terminated with the error.
"Fortran runtime error: Index '301' of dimension 2 of array 'dmap' above upper bound of 300".
A. This error was caused by the limitation of memory space in MAINMAST program. In order to reduce the size of map file, we can use "relion_image_handler" command. The size of map should be smaller than 15-*150*150.
Sample command: Shift map to their center-of-mass and change voxel size to 100*100*100.
relion_image_handler --i INPUT.mrc --new_box 100 --o OUTPUT.mrc --shift_com true
Q. I do not have the relion program in my PC. How can I reduce the size of EM map?
A. If you have a reference pdb file. You can extract the regions within a specific distance of the currently selected atoms (or chains/molecules) by "zone tool" in UCFS Chimera. See Here
- Select chain A:
[Chimera menu] select > Chain > A
- Segment out around chain A:
[Volume Viewer Menu] Features>Zone
Click [Zone] botton.
- Save the segmented map:
[Volume Viewer Menu] File>Save Map as...
[Chimera menu] select > Chain > A
[Volume Viewer Menu] Features>Zone
Click [Zone] botton.
[Volume Viewer Menu] File>Save Map as...
Parallel Computing
Q. Can I calculate MAINMAST modeling on cluster computers?
A. Yes. You can use MAINMAST program with slurm job submission system on your computer clusters.
We are providing our python script that generate job files for slurm's sbatch command.
Plese check MAINMAST website Section:MAINMAST program on cluster machines HERE
Paramaters
Q. How important is the map density threshold? Can I use 1.0 (or 0.9, 0.8, ...)*recommended contour level ?
A. If the main-chain structure is clearly observed at high contour level, we should not use low threshold values. Because using low density threshold will increase the number of the local dens points and noise data. It directly affects to the computational time.
Q. How can I find the best density threshold for my the EM map?
A. Before calculating the main-chain models, you can inspect the initial Minimum Spanning Tree (MST) structure. If the MST does not cover many regions of MAP, the threshold value is too high.
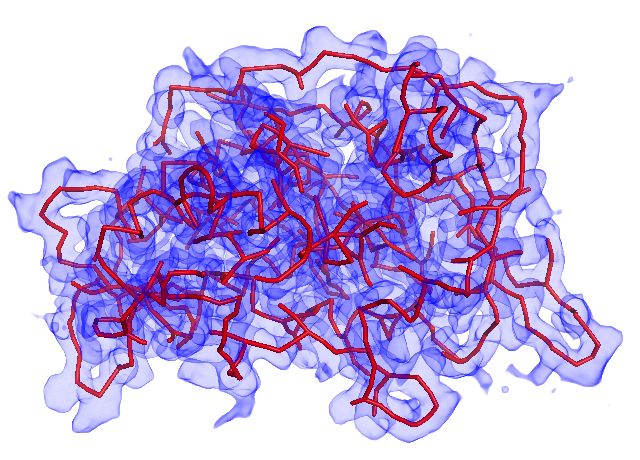
MST with threshold=1.0
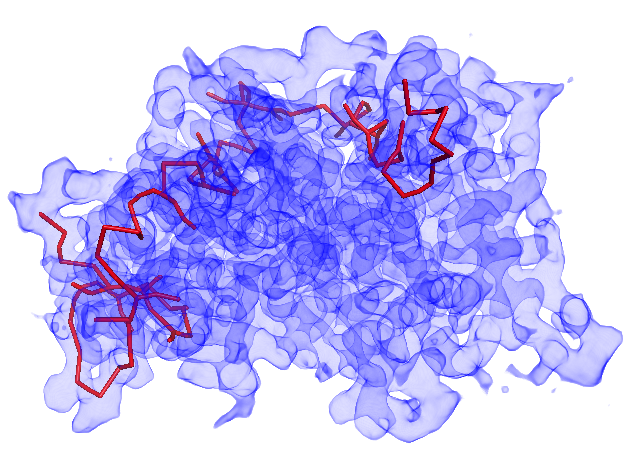
MST with threshold=8.0. Many missing regions.
© 2026 KIHARA Bioinformatics LABORATORY, PURDUE University | Design by TEMPLATED.
