
| Home | Server | LZerD | Multi-LZerD | PI-LZerD | IDP-LZerD | Path-LZerD | Flex-LZerD | About |

|
|||||||||||||||||||
|
|
PI-LZerD: Protein Docking Prediction Using Predicted Protein-Protein Interface PI-LZerD uses protein interface predictions from current protein interface prediction methods along with our pairwise protein-protein docking program, LZerD, iteratively to reach higher prediction accuracies. The suggested citation when referring to PI-LZerD is: Protein docking prediction using predicted Protein-Protein Interface, Bin Li, Daisuke Kihara, BMC Bioinformatics, 13:7 (2012). The following is a summary diagram of PI-LZerD: 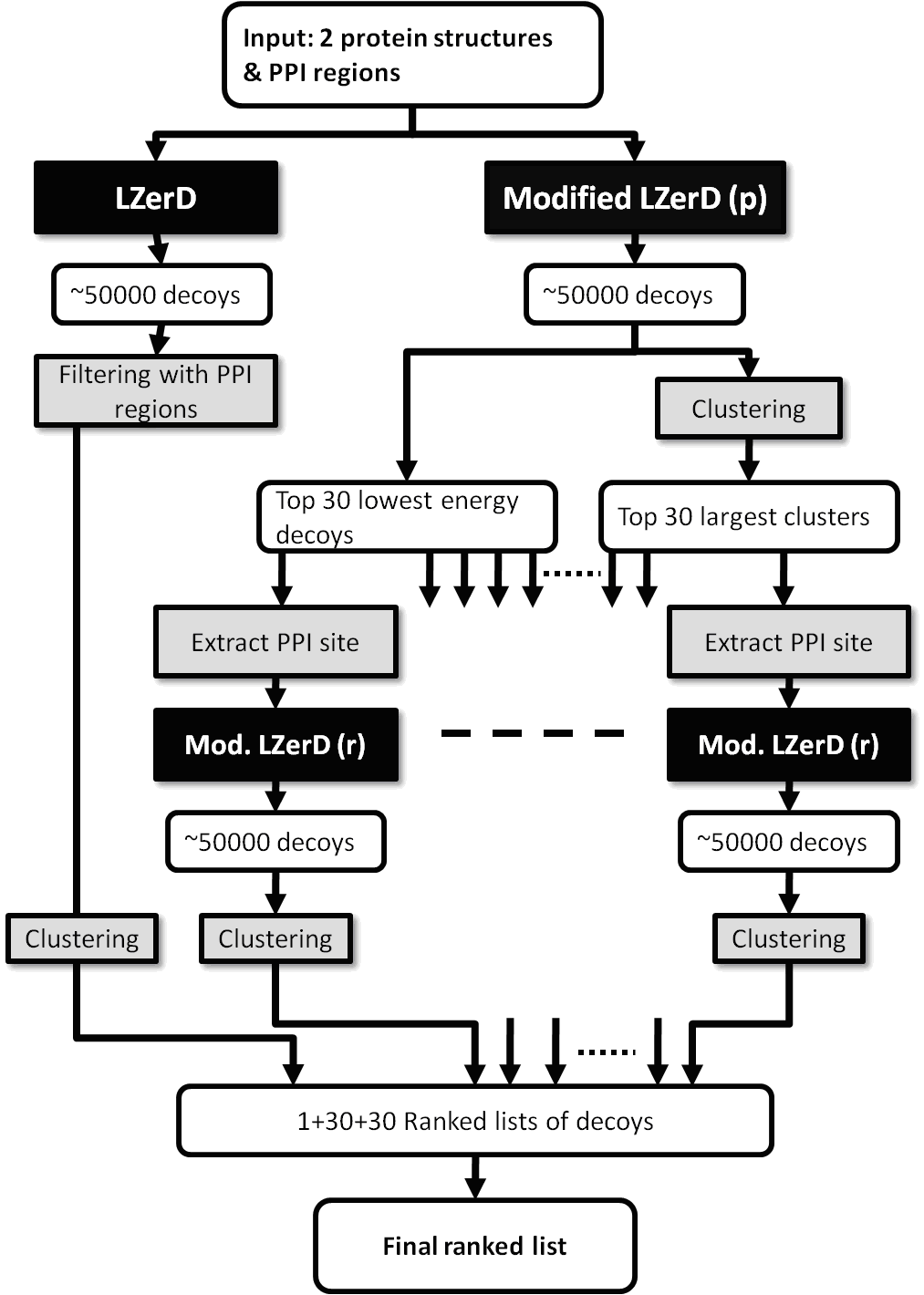
Dependence As shown in the following figure, PI-LZerD process includes five major steps. For each major step, many sub-steps are stored into seperated directories. Following graph shows an overview of the major steps, as well as the dependence of each individual steps.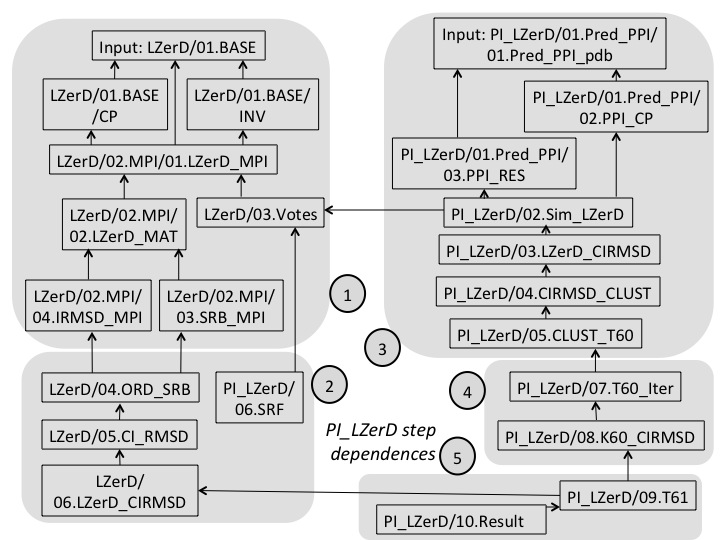
Resources The 124 PDB files of bound and unbound datasets used. The residue indices of correct, 5 residues shift, 10 residues shift, 12 residues shift, 15 residues shift for bound cases. The PDB files used when meta-PPISP was used as basis for the PPI predictions. Copyright Copyright of this distribution belongs to Bin Li, Daisuke Kihara. It's free for academic non-profit institutions. For commercial entities or government research labs, please contact us (dkihara@purdue.edu) to get the approval to use this distribution. Redistribution of any files in this package without our permission is prohibited. Download PI-LZerD The binary executable files can be downloaded here. Installation instructions are given in the README file provided. FEEDBACK If you have any questions or suggestions, please feel free to contact us (dkihara@purdue.edu). This package will be updated based on user feedback. |
| Copyright © 2026 KIHARA Bioinformatics LABORATORY, PURDUE University | |