| File | About |
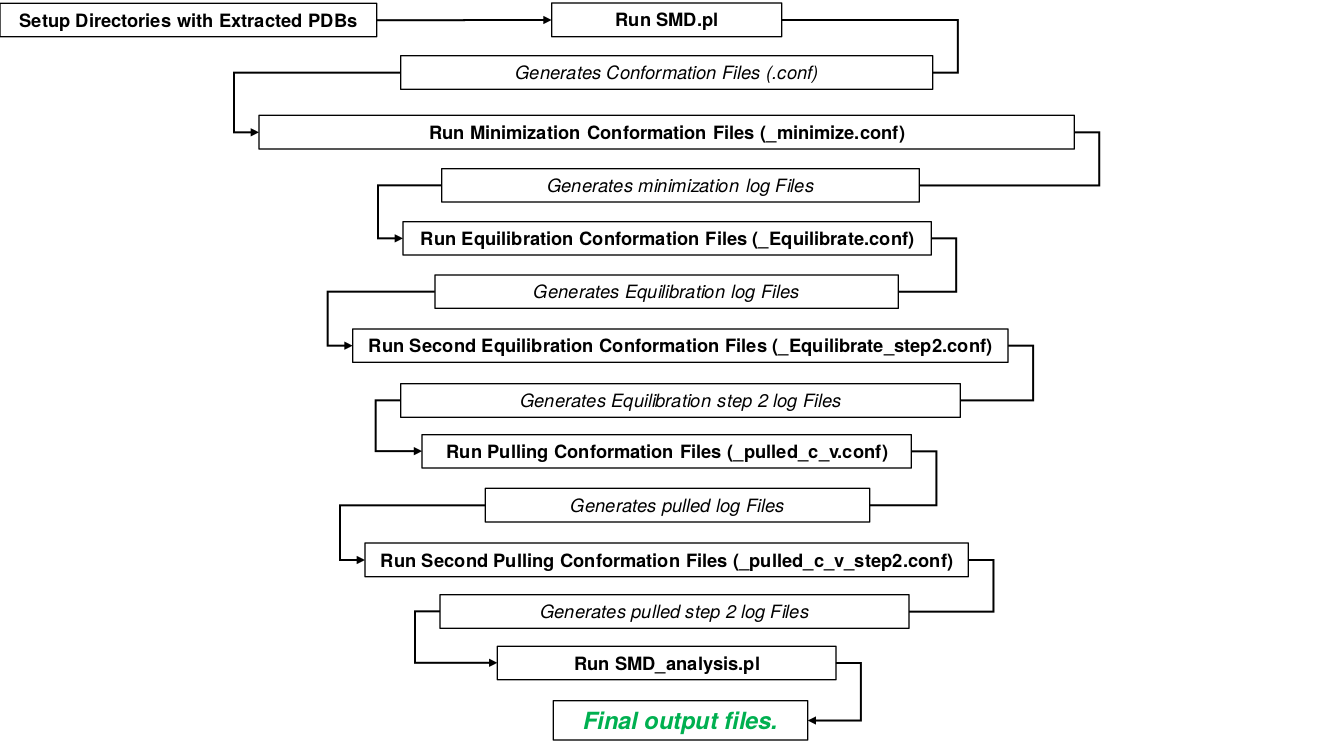
| pdb.tar | PDB data set of casp models used in the study, organized by target number. There are a total of 36 target sets with 2976 models in total. Models have names T0###TS###_#.pdb, where # is an integer between 0 and 9. Once untared, run SMD.pl, then run namd simulations. |
| SMD.pl | Perl script for setting up NAMD steered molecular dynamics simulations. Runs on all .pdb files in a directory called ./pdb/ |
| SMD.pm | Subroutines for SMD.pl |
| SMD_analysis.pl | Perl script for the analysis of log files from SMD simulations. Formats force-time curves and finds Peak data. |
| SMD_analysis.pm | Subroutines for SMD_analysis.pl |
| README.txt | How to set up and run the perl scripts. |
| parameter files | |
| par_all36_prot.prm | CHARMM36 parameter file for proteins used. |
| toppar_water_ions_namd.str | Parameter file for water and ions. |
| Example Output |
| minimize.log | An example minimization output log file. This is the output from running namd2 model_minimize.conf as decribed in the README. |
| Equilibrate.log | An example equilibration output log file. This is the output from running namd2 model_Equilibrate.conf as decribed in the README. |
| Equilibrate_step2.log | An example step 2 equilibration output log file. This is the output from running namd2 model_Equilibrate_step2.conf as decribed in the README. |
| pulled_c_v.log | An example pulling output log file. This is the output from running namd2 model_pulled_c_v.conf as decribed in the README. |
| pulled_c_v_step2.log | An example step2 pulling output log file. This is the output from running namd2 model_pulled_c_v_step2.conf as decribed in the README. |
| Force.dat | An example force curve. Plot data with gnuplot or excel. This kind of file is generated by SMD_analysis.pl. |
| Force_smooth.dat | An example smoothed force curve. Plot data with gnuplot or excel. This kind of file is generated by SMD_analysis.pl. This is the data that is used to determine the break force. |
| Force_smooth.dat | An example output of the Model GDT_TS and Force from SMD_analysis.pl |