|
Introduction
The steady increase in the number of electron microscopy
maps deposited in the EMDataBank
requires the development of search tools that can compare
the isosurface of maps to help understand the
relationships between them.
EM-SURFER provides a web-based infrastructure to
rapidly compare 3D EM maps based on isosurfaces derived from
author-recommended contour levels.
The features provided to users are discussed in detail below.
Integrative Web Interface:
EM-Surfer provides an easy-to-use search interface
where a target EM map's isosurface shape is compared against
the EMDataBank, using 3D Zernike Descriptors as an efficient
shape descriptor. The query provided by the user can
be compared against several surface representations. It
can take into account volume differences between the target
and maps in the database. Results can be easily
visualized in a user-friendly search results page. By
clicking one of the maps retrieved, a subsequent search
is invoked using the map clicked as a query to further
navigate 3D EM maps.
3D Zernike Descriptors
3D Zernike Descriptors (3DZD) are utilized for the
efficient comparison of EM map isosurfaces. The descriptor
is a combination of coefficients calculated from a
set of orthogonal 3D basis polynomials that
approximate a given 3D function (e.g. a grid representing
the EM map). 3DZD has various desirable properties when
applied to EM maps:
- Rotational invariance: Prior
structural alignment is not required for map
comparisons.
- Compactness: An individual surface can
be compactly represented as a feature vector with only
121 numbers (called invariants). Comparisons of these
vectors can be performed by calculating the
Euclidean distance in a speedy fashion,
thus enabling rapid shape retrieval. Furthermore,
the concatenation of vectors that describe the same
map at different contour levels create richer
descriptors (up to 242 and 363 numbers in
EM-SURFER).
- Hierarchical Resolution: Invariants
of lower resolution are also part of the higher
resolution. For example, the first 12 numbers
among the 121 invariants represent a coarse-grained
version of the surface, while the complete 121
invariants provide the best approximation of
the surface that the polynomials can give us.
For more technical details about the use of the 3DZD for EM map comparison, please refer to our previous work
[Sael L. and Kihara D. , 2010]
.
3DZD Computation Procedure
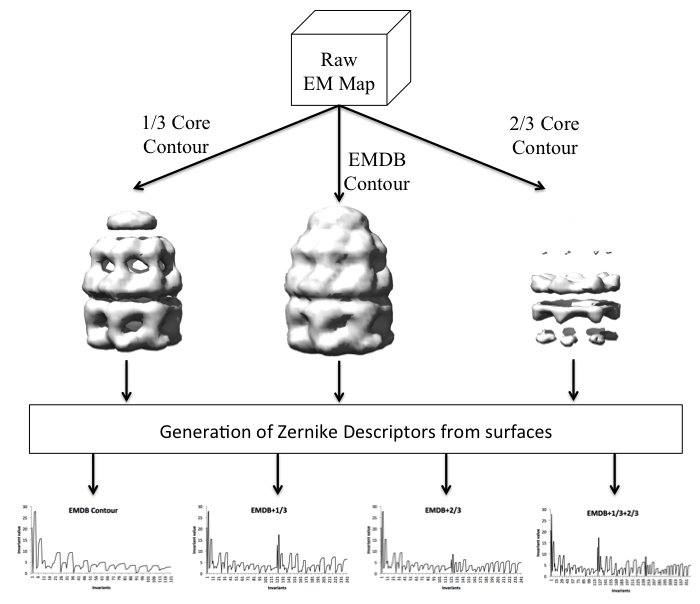
- Isosurface generation:
Three isosurfaces at different density levels are
created for each EM map in the database.
One comes from the
author-recommended contour, as specified in the
EMDataBank. Additionally, two higher isovalues
are used to represent regions that are closer to
the core of the molecules.
In 1/3 Core Contour, an increased contour level,
1/3 * (max density - recommended contour) is used.
The same idea applies for 2/3 Core Contour, just
changing the factor from 1/3 to 2/3.
- 3D Zernike transformation:
The 3DZD program [Novotni M. and Klein R, 2003]
takes the cubic grid as input and generates
3DZDs for each surface representation
(121 invariants).
- Zernike invariant combinations:
The previous step creates three vectors of size 121.
The various shape representation
alternatives offered by EM-SURFER arise from a
combination of these. For example,
EMDB contour+1/3+2/3 uses a descriptor
vector of size 363, by concatenating all three
vectors. On the other hand, EMDB contour
uses only the 121 invariants from the
author-recommended contour value.
Search Results by EM-SURFER
A search against the whole EMDB entries can be performed from the search page .
- In Step 1, choose the contour shape representation. The default is set to the author-recommended contour level, but users can choose any of the other 3 options.
- In Step 2, choose the EMDB entry ID or upload an EM map file. To find an ID by a text search using for example, protein name, use the
EMDB search page.
Upload troubleshooting: While EM-SURFER has several checks and repair machanisms to process uploaded files, it is possible that some errors arise. These situations occur because of network connectivity problems while uploading files, incorrect formatting in the EM file uploaded, etc. If you experience a problem uploading your map please contact dkihara@purdue.edu
- In Step 3, a volume filter is provided. The default is on. When this filter is on, a search only retrieves EM maps that have similar volume to the query.
The figure below explains the search result page:
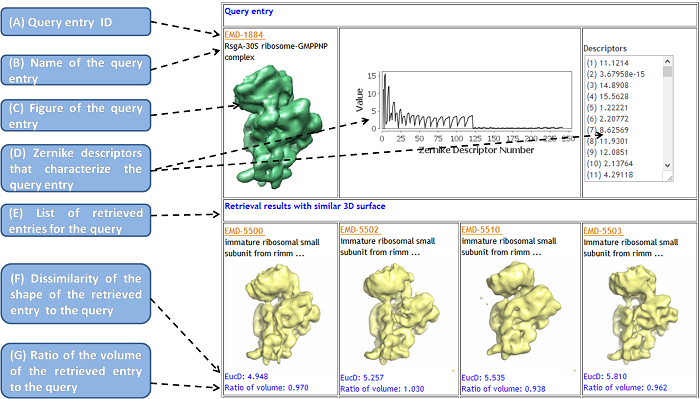
- Query entry ID
- It is a unique 4-digit accession number for each EMDB entry, which can also start along with "EMD-".
- Name of the query entry
- The description information about the entry is extracted from the XML file of the entry in the EMDB. If the length of the characters of the entry is larger than 40, the initial 40 characters and ellipsis (...) are shown, and the full information is presented on the popup box.
- Figure of the query entry
- A figure of an isosurface of the entry is shown. It is provided from the EMDB.
- Zernike descriptors that characterize the query entry
- The Zernike Invariants (or Zernike Descriptors), which characterize each EM isosurface are displayed in text and a graphic form.
- By clicking on the figure of retrieved entries, a new search will be invoked against the whole EMDB from the clicked entry.
- List of retrieved entries for the query
- When clicking on the figure, it can smoothly invoke the iterative search of the similar EM isosurface on the target entry.
- The retrieved EM Surfaces from the database are shown on the panel below. They are ranked by their Euclidean distance of the 3DZDs to that of the query entry. In default setting, the top 20 similar surfaces are displayed.
- Dissimilarity of the shape of the retrieved entry to the query
- The dissimilarity of two EM isosurfaces are quantified by
the Euclidean distance (the square root of the sum of the squares
of the differences between corresponding values) between the 3DZD vectors of the retrieved isosurface against the query.
-
The smaller the EucD value, the more similar the shape of the two EM map isosurface.
-
Empirically, related entries have an Euclidean distance of less than 10.0.
- Ratio of the volume of the retrieved entry to the query
-
The ratio is defined as the volume of the retrieved EM entry in the database devided by the volume of the query entry.
Job Submission and 3DZD Computation in Batch Mode
When users would like to benchmark EM-SURFER by submitting a large number of queries,
they can use the batch mode page .
-
When submitting a batch of queries to EM-SURFER,
users can either type a custom list of EMDB IDs or upload a separate file with those IDs.
-
Taking the same steps as the submission of a single entry, users can go through the page to select an isosurface representation, and specify the volume filter.
-
The extra step in this section is to select the number/size of the retrieved list for each query.
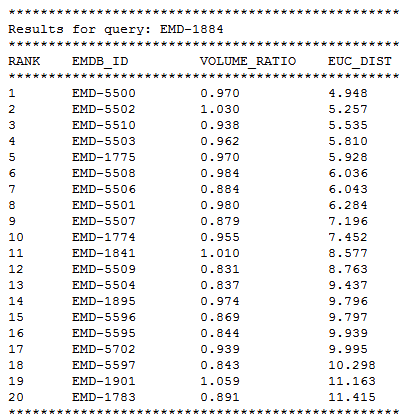
Taking EMD-1884 as an example, The figure below shows its retrieved result list of the top 20 most similar isosurface in EMDB.
When users would like to obtain 3DZD of a large number of queries, they can also use the
batch mode page .
-
After selecting isosurface representation in "Step 1", query proteins are then provided by
either entering their ID codes in "Structure id list" box or by uploading a structure id list
file in "Step 2". Users can click on "Get 3D Zernike Descriptor" button in "Step 3" to open 3DZD
result page in a new tab.
References
- Lee Sael and Daisuke Kihara, Protein surface representation for application to comparing low-resolution protein structure data.ABMC Bioinformatics 2010;11:S2.
- Novotni M, Klein R. 3D Zernike descriptors for content
based shape retrieval.ACM Symposium on Solid and
Physical Modeling, Proceedings of the eighth ACM
symposium on Solid modeling and Applications 2003;216-225.
|